Research Projects
Project 1: Regulation of flight muscle specific splicing by Bruno1
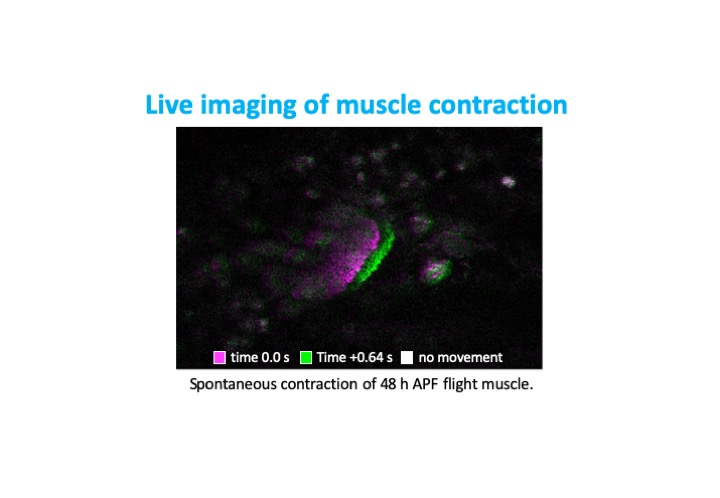
Bruno1 (also called Bru1, Arrest, Aret) is a conserved member of the CELF-family of RNA binding proteins containing 3 RRM domains. The human homolog CELF1 is known to be mis-regulated in myotonic dystrophy, and together with MBNL to control many of the RNA regulatory processes disrupted in patient muscles. I previously showed that Bru1 regulates flight muscle specific alternative splicing in Drosophila, and its loss leads to defects in sarcomere growth, hypercontraction and ultimately muscle fiber loss. In a project funded by the Deutsche Forschungsgemeinschaft, we are investigating the mechanism by which Bru1 regulates splicing in flight muscle.
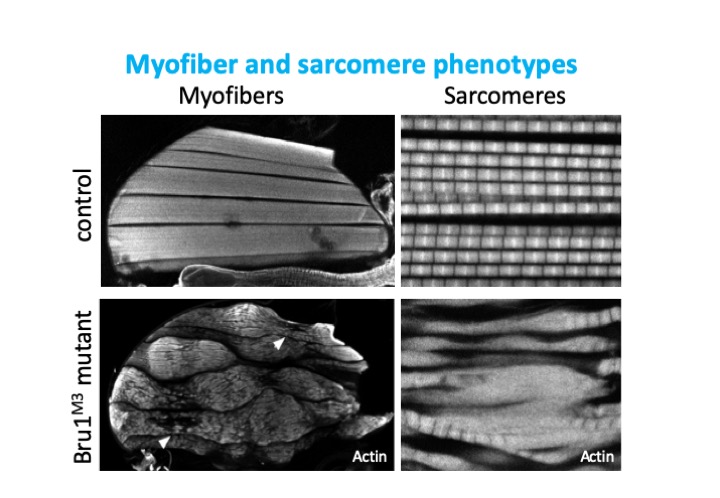
To understand developmental mechanisms leading to myofibril phenotypes in Bru1 mutants, we use genetics, confocal microscopy and transmission electron microscopy. We are further evaluating the function of different Bru1 isoforms, including an uncharacterized isoform containing only 2 RRMs. We also would like to identify Bru1 binding elements in direct RNA targets using the iCLIP technique. We expect to define regulatory elements in introns or exons near splice junctions regulating alternative splicing and possibly in UTR regions regulating mRNA localization, stability or translation. These studies will identify the developmental mechanisms misregulated in Bru1 mutants leading to defects in myofibril growth, assign specific functions to individual Bru1 isoforms and identify direct Bru1 targets in flight muscle. These findings may have vertebrate relevant implications, in particular for the function of CELF family members in muscle development and disease.
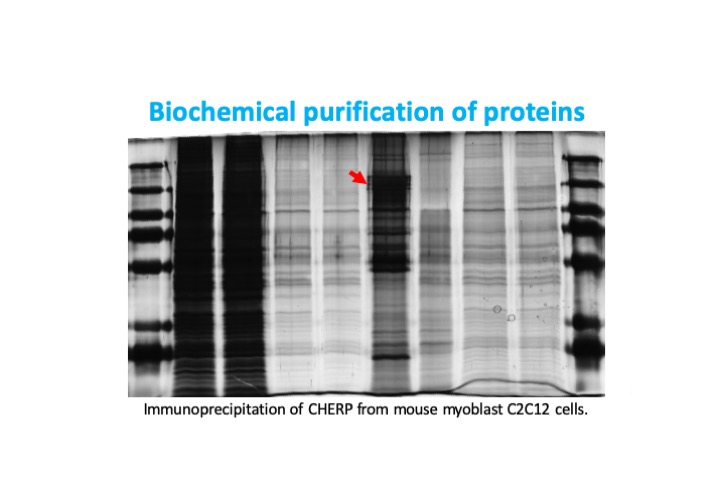
Project 2: Scaf6/DmCHERP function during flight muscle development
CHERP (Calcium homeostasis endoplasmic reticulum protein) was first identified in vertebrate cell culture to play a role in cellular calcium homeostasis, but has since been shown to be an accessory component of the U2 spliceosomal complex. Together with Rbm17 and U2SURP it controls alternative splicing as well as suppressing cryptic splice sites. In certain cancers, its overexpression is sufficient to drive proliferation and suppress apoptosis, suggesting it can act as an oncogene. Our preliminary data implicates the Drosophila homolog of CHERP, Scaf6, in muscle development.
In collaboration with Dr. Rippei Hayashi from the Australian National University, we are working to understand Scaf6 function in Drosophila development. We are focused on characterizing mutant and RNAi knockdown phenotypes in muscle and other fly tissues. We hope to gain detailed mechanistic insight into Scaf6 function through RNA-Seq and mass spectrometry experiments. We are further investigating the evolutionary conservation of Scaf6/CHERP function in mouse myoblast C2C12 cells. Our results will provide insight into spliceosome function and the coordination of different splicing processes with relevance to myogenesis and the regulation of cell proliferation.
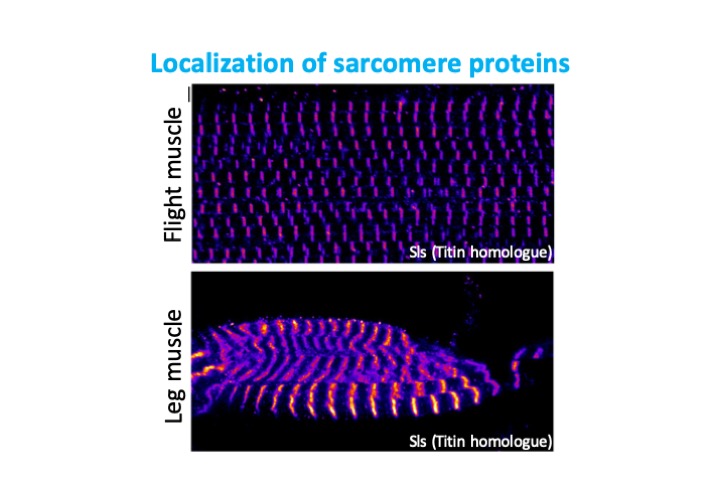
Project 3: Identification of RNA binding proteins in muscle
There are hundreds of proteins known to bind RNA, mostly containing one or more canonical RNA-binding domains. However, recent RNA-interactome capture studies have demonstrated that far more proteins than previously appreciated have the ability to bind RNA. The function of this binding is currently poorly characterized, as is the tissue-specificity of the proteins shown to bind RNA. We are interested in identifying proteins that bind RNA in muscle, potentially impacting alternative splicing, transport, stability, translation and other steps in RNA regulation in a cell-type specific manner.
We employ several different approaches to identify new RNA binding proteins and characterize their function in muscle. We identify tissue-specific candidates from genome-wide deep sequencing and RNAi screen datasets, as well as using biochemical purification and omics technologies. We further plan to develop fluorescent reporters for defined muscle-type specific alternative splice events and use these to screen for factors controlling a define set of alternative splice events, harnessing the power of forward genetics in Drosophila. Our data will identify novel RNA binding proteins, new pathways regulating RNA metabolism and tissue-type specific logic in splicing regulation.