Research Projects
Eukaryotic genomes are organized into distinct functional domains that are transcriptionally active (euchromatin) or repressed (heterochromatin). However, while many of the components and enzymes contributing to the establishment and spreading of heterochromatin have been identified, we still have a poor knowledge of the spatiotemporal control of this process: What favors the formation of heterochromatin? Which factors contribute to these processes? What are the underlying mechanisms? What limits its spreading beyond natural boundaries? How are these pathways coordinated? How are they dynamically regulated?
To address these questions, our lab applies several strategies. Using functional genome-wide screens, we seek to identify novel factors that modulate the heterochromatic state. Using functional genomics, we examine how these factors cooperate by dissecting regulatory pathways and networks. Using live-cell imaging, molecular biology and biochemistry methods, we investigate the mechanism of heterochromatin regulation by these factors.
1.) Identifying novel factors and dissecting regulatory networks
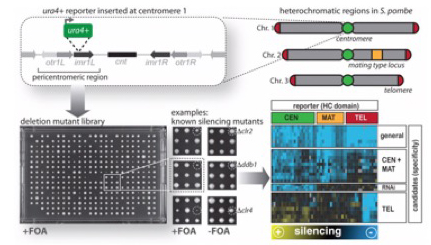
Through reporter-based genetic screens, we have identified a large number of factors that modulate the silent state of heterochromatin domains. To understand the roles of these novel factors in heterochromatin regulation, we analyze their genetic interactions quantitatively and at the genome-wide level using SGA (Synthetic Gene Arrays). This allows us to dissect mutations within the same pathway displaying non-additive phenotypes from mutants of parallel pathways exhibiting synthetic phenotypes (Verrier et al. Open Biol 2015; Barrales et al. Genes Dev 2016; Flury et al. Mol Cell 2017; Salas-Pino et al. JCB 2017). Future work aims on assessing systematically the pair-wise genetic interactions of all identified factors by an advanced version of the E-MAP (Epistasis Mini-Array Profile) approach and examining the role of stress in heterochromatin regulation. [Read more]
2.) Demarcation and protection of heterochromatin domains
|
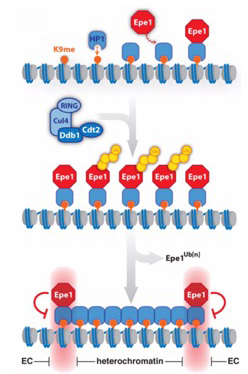
Partitioning into active and silent chromatin requires mechanisms that specify boundaries and maintain the identity of chromatin domains. We previously discovered a novel mechanism by which the chromatin distribution of an anti-silencing factor is shaped through ubiquitin-dependent degradation that we coined ‘chromatin sculpting‘ (Braun et al. Cell 2011). The integrity of chromatin domains can also be protected through anchoring to specific chromatin sites, thereby preventing promiscuous binding. In collaboration with Marc Bühler’s lab, we showed that the methyl-reader Pdp3 recruits the acetyltransferase Mst2 to euchromatin via recognition of methylated H3K36 to prevent ectopic heterochromatin and mistargeting of Mst2 to heterochromatin triggering a silencing defect (Flury et al. Mol Cell 2017; Georgescu et al., Microbial Cell 2020). [Read more] |
 |
3.) Regulation of gene repression at the nuclear periphery
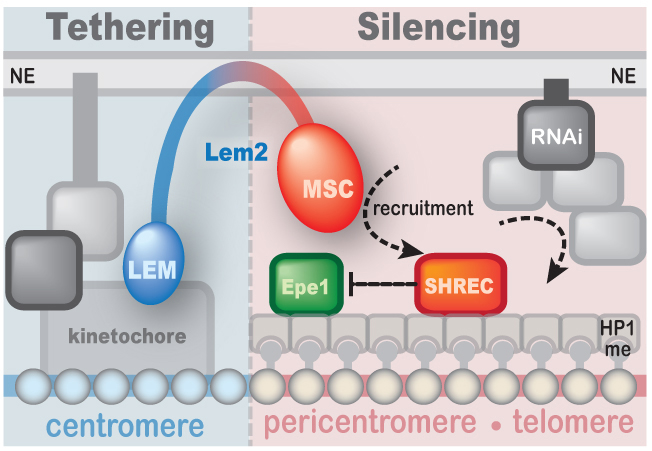
The nuclear periphery provides a specialized subcompartment that promotes the establishment and maintenance of silent chromatin. We showed that the inner nuclear membrane protein Lem2 contributes to silencing and localization of heterochromatin and is part of a network of multiple redundant silencing pathways (Barrales et al. Genes Dev 2016). While Lem2 associates with centromeric chromatin via its LEM domain, it mediates silencing through its conserved MSC domain. Thus, centromere localization and silencing can be mechanistically separated, revealing an unanticipated complexity [Read more]
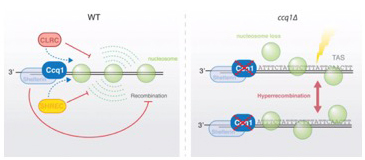
4.) Regulation of repetitive sequences and genome maintenance
DNA repeats are often embedded into heterochromatin to prevent unwarranted recombination and genome instability. However, during DNA damage or loss, recombination of these repeats may become necessary. We showed recently that repeat-like sequences present at subtelomeres display metastable nucleosomes and are hyper-recombinogenic, promoting telomere plasticity and recombination (van Emden, Forn et al., 2019). This is an intrinsic property of the underlying DNA sequences and counteracted by Ccq1, a member of the conserved shelterin complex. We are currently testing whether these fragile DNA sequences play an active role in telomere maintenance independent of telomerase.