Marta Russo
Our group will move to the BMC in May 2025.
Our lab investigates how metabolism influences the regulation of gene expression, with a particular focus on the nuclear compartment. While metabolic pathways have traditionally been studied in mitochondria and cytosol, there is growing evidence that key metabolic enzymes also localize to the nucleus, where they may directly fuel epigenetic and transcriptional processes. Understanding how these nuclear metabolic activities shape chromatin structure and gene regulation is central to our research.
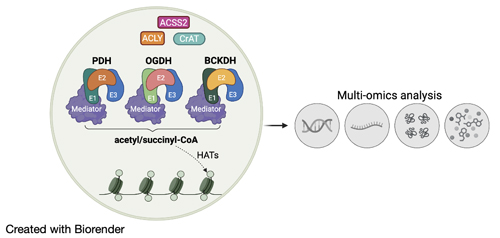
We focus on the local production of acyl-CoA metabolites in the nucleus and their role in regulating gene activity. In particular, we study nuclear-localized 2-ketoacid dehydrogenases—namely pyruvate dehydrogenase (PDH), oxoglutarate dehydrogenase (OGDH), and branched-chain α-ketoacid dehydrogenase (BCKDH)—as active producers of acetyl-CoA and succinyl-CoA within the nuclear environment.
Our recent work revealed that these metabolic enzyme complexes physically associate with the Mediator complex, a central coactivator of RNA Polymerase II-dependent transcription, and remain catalytically active at chromatin. We showed that this local metabolic activity supports histone acetylation, linking the metabolic state to epigenetic regulation at specific genomic loci.
We use a multidisciplinary approach that combines genomics (ChIP-seq, RNA-seq), mass spectrometry-based proteomics, metabolite tracing, and structural biology techniques (including XL-MS and AlphaFold modeling) to explore:
• how nuclear acyl-CoA-producing enzymes regulate transcription during immune activation,
• how they interact with chromatin-associated factors beyond Mediator,
• and how their structural organization and subnuclear localization support specialized regulatory functions.
Our primary model system is the macrophage, an immune cell type that undergoes rapid transcriptional and metabolic reprogramming during inflammation. However, we also investigate how these mechanisms extend to other mammalian cell types, with broader implications for understanding immunometabolism, development, and disease states.