Maria Spletter
RNA-dependent regulation during development processes
https://spletterlab.de
For Non-scientists:
We rely on our muscles for simple movements, from picking up a pencil to walking across a room. Muscles also enable unconscious movements, such as beating of the heart, the digestive action of the stomach or breathing. Muscles are able to facilitate movement because they contract, meaning they are able to shorten and reextend. This contraction ability is powered by mini-motors called sarcomeres.
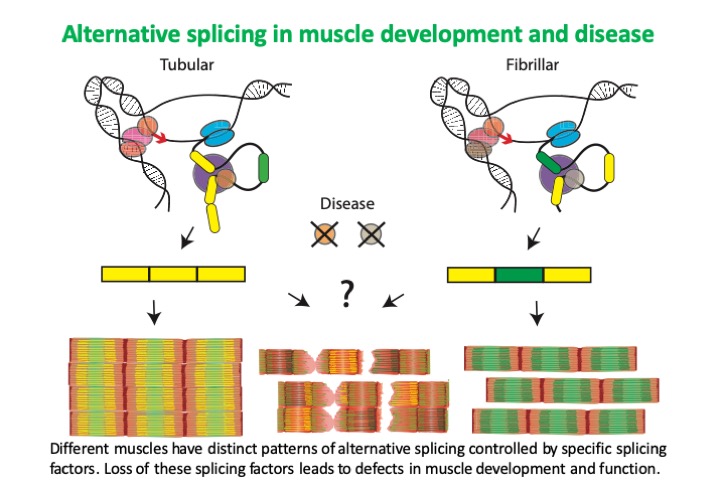
We have hundreds of muscles in our bodies with different functions and distinct contractile properties. To achieve this, muscles modify their sarcomeres to adapt to physical demand. For example, the heart needs to be very stiff to properly pump blood, while muscles in the legs are more flexible to permit walking. In addition to regulating which proteins are present to build their sarcomeres, muscles also produce different versions of the same protein, termed isoforms, using a process called alternative splicing. Alternative splicing generates multiple versions of the same protein that for example can be more or less stiff or interact more strongly or weakly with other proteins, ultimately helping define specific contractile properties for different muscle types.
There are many regulators that help a muscle select which proteins and protein isoforms it will produce. Misregulation or loss of regulators responsible for alternative splicing causes muscle diseases such as myotonic dystrophy and dilated cardiomyopathy. These disorders cause muscles to produce the wrong protein isoforms, for example a version of the protein normally found in leg muscle can be found in heart. Patients with these disorders experience a loss of muscle and impaired muscle function, dramatically decreasing their quality of life and life expectancy. The goal of our research is to understand how regulators of alternative splicing work in muscle, and what goes wrong in muscle disease. A better understanding of how muscles develop normally and what is disrupted in disease will aid in developing new treatments for these debilitating disorders.
For more information, please see Dr. Spletter’s 2018 TEDxTUM talk on “What fruit flies can tell us about muscle disease.” https://www.youtube.com/watch?v=Uhg75v5_Xu8

For Scientists:
In our research we strive to understand the normal function and regulation of RNA processing, in particular alternative splicing, in myogenesis and how dysregulation contributes to muscle disease. After transcription, RNAs have to be spliced, capped and poly-adenylated, edited, exported from the nucleus, trafficked and translated. Each of these steps is heavily regulated, and additional pathways impact transcript stability. Regulation is achieved largely through the activity of RNA binding proteins, canonically identified to have at least one of a panel of characterized RNA binding domains. These processing steps determine the mRNAs that are available for translation, determining which proteins and protein isoforms are ultimately expressed by a cell.
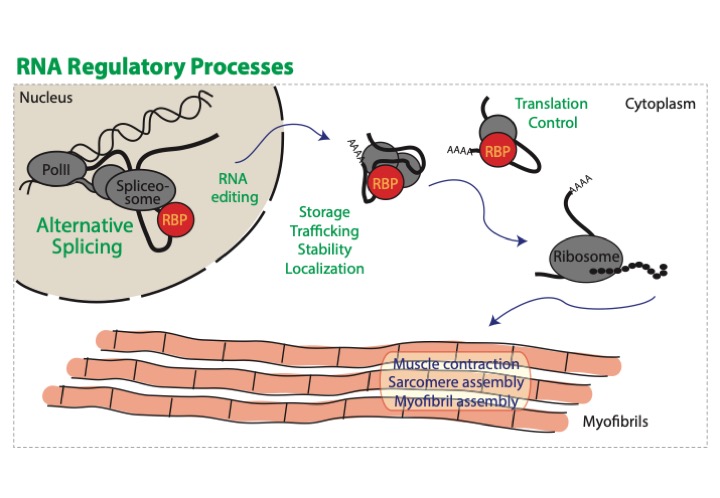
Muscles rely on alternative splicing and RNA regulation to fine-tune their contractile properties. Expression of different isoforms of sarcomeric proteins, for example myosin, actin or titin, have been demonstrated to impact actomyosin contractility. During normal development and in response to exercise, different muscles display distinct patterns of splicing and protein isoform expression. These expression patterns are frequently disrupted in disease, resulting in expression of the wrong sarcomere protein isoforms and leading to structural defects, disruption of function, atrophy and in many cases fiber loss. Diseases such as dilated cardiomyopathy and myotonic dystrophy are caused by dysregulation of RNA binding proteins, and many other diseases are characterized by misregulation of splicing or RNA trafficking. Despite this, our understanding of RNA regulatory mechanisms as well as the proteins capable of modifying and regulating RNA is limited and incomplete.
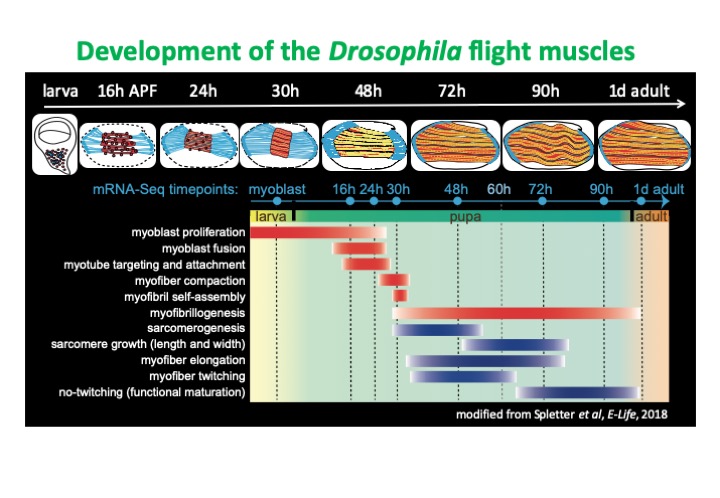
We investigate RNA regulation and muscle development using mainly Drosophila melanogaster as our model organism. In addition to the vast array of genetic tools and fast generation time, muscle proteins and structure are highly conserved from flies to vertebrates. In particular we focus on the indirect flight muscles (IFMs), which are distinct from other fly muscles with their asynchronous and stretch-activated contractions and lack of lateral alignment between myofibrils. The development of the IFMs is well characterized and stereotyped, allowing detailed investigation of basic mechanisms of sarcomere formation, myofibril organization and muscle maturation. Using this model, we hope to understand the mechanisms guiding tissue-specific alternative splicing and RNA regulation, as well as identify novel splicing regulators. Our results in Drosophila will be directly relevant to understanding mechanisms of pathogenesis in human muscle disease, as well as conserved mechanisms of muscle development and sarcomerogenesis.