Research Projects
1.) Regulation of transcription factor function by metabolites and molecular glues
Obesity and insulin resistance have become a pandemic. 20% of Europeans will develop type 2 diabetes (T2D) by 2030. Carbohydrates such as glucose, fructose and ethanol contribute to the metabolic syndrome. They activate gene programs that convert excess carbohydrates into fat. How this happens is not well understood at the mechanistic level. Since current therapies fall short of medical needs, more insights into how nutrients regulate gene activity directly is required.
Our team has discovered the 1st cellular receptor for an important cellular glucose metabolite. The endogenous metabolite and specific chemical analogs act as high-affinity molecular switches (agonists and antagonists) of a conserved human transcription factor. This factor is essential to control sugar and lipid homeostasis and also regulates insulin sensitivity as well food intake. We are now pursuing the molecular analysis of several transcription factors that are targeted by cellular metabolites and whose activity we will influence using synthetic small molecules. This set of projects has great potential to translate into new cures for human metabolic syndromes and also be highly relevant intreating several cancers, where related molecular interactions drive altered protein function and are thus amenable through chemical entities. This project is in part funded by a recent BioM m4 award.
2.) Mechanistic insight into genome-maintenance catalysed by the histone chaperone FACT
The multisubunit FACT (Facilitates Chromatin Transcription) complex, which is composed of the subunits Spt16 and SSRP1 (encoded by Pob3 and Nhp6 in fungi), is the only essential histone chaperone in S. cerevisiae. It facilitates RNA polymerase II-mediated transcriptional elongation and DNA replication, promotes histone dynamics and is vitally required for the proper assembly of chromatin structure. Over the last few years, we discovered that the Pob3 subunit is required to maintain a fully functional heterochromatin structure at fission yeast centromeres. We identified and characterized a histone H3-H4 binding function residing in the N-terminal aminopeptidase-like fold of the Spt16 subunit and solved its high-resolution crystal structure. We are now comprehensively dissecting the histone chaperoning functions of the FACT complex. In the future, we plan to extend our work to obtain the high-resolution structure of the complex between FACT and histones H2A-H2B.
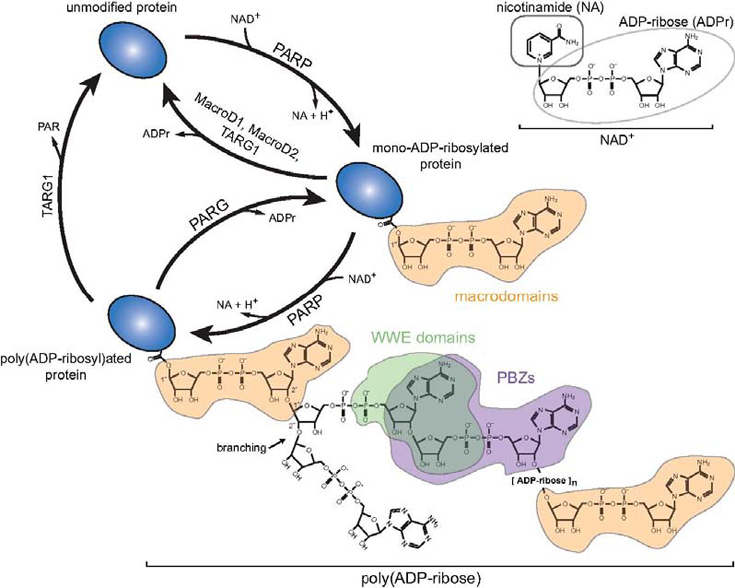
3.) Molecular dissection of ADP-ribosylation signalling
We are interested in deciphering the biological role and cell biology of ADP-ribosylation, a highly dynamic and reversible post-translational modification with a long history, clear clinical importance, yet little mechanistic insight. This modification is actively involved in regulating chromatin structure and transcription upon a variety of environmental stresses, most notably DNA damage. DNA damage events require attention by the nuclear repair machinery or cells will accumulate mutations that may result in cancer.
We are studying how the enzymatic activity of the DNA damage-activated ADP-ribosyltransferase PARP1 is regulated by the recognition of broken DNA strands. The precise molecular mechanism through which PARP1 activity is strictly controlled by the recognition of damaged DNA remains unknown. We are investigating the functional requirements for DNA binding under damage and non-damage conditions in vivo using life-cell imaging of PARP1 enrichment at DNA damage sites, including FRAP- and FCS-based approaches. We study how the enzymatic activity of the ribosyltransferase PARP1 and the ATP-dependent nucleosome remodeler Alc1/Chd1L are strictly regulated by their recognition of damaged DNA in life cells. This has allowed us to identify the first set of globular protein modules capable of directly recognizing poly-ADP-ribose (the so-called macrodomains).
This discovery now allows us to study a whole family of proteins that respond to PARP signalling. Biochemical evidence shows that the enzymatic activity of the macrodomain-containing Alc1/Chd1L enzyme, for example, fully depends on PARP1 activation. In addition, we are dissecting how Alc1/Chd1L and other nucleosome remodelers recruit to DNA breaks.